Machine Learning to Fight Antimicrobial Resistance
Antimicrobial Resistance (AMR) occurs when bacteria, viruses, fungi and parasites change over time and no longer respond to medicine. Seven key machine learning projects to fight against antimicrobial resistance are discussed. Artificial Intelligence (AI) is also used to hunt the new resistance genes, for building better understanding of how bacteria fight off drug treatments.
Antimicrobial resistance is one of the key reasons for human sufferings in modern hospitals. The objective of our Compassionate AI Lab research is to eliminate the pain and sufferings of the humanity with the use of emerging technologies. Overcoming the challenges of antimicrobial resistance is one of the key research area of our Compassionate AI Lab.
Preventing microbes from developing resistance to drugs has become as important issue for treating illnesses across the world. Artificial Intelligence, machine learning, genomics and multi-omics data integration are the fast-growing emerging technologies to counter antimicrobial resistance problems. Here, Dr. Amit Ray explains how these technologies can be used in seven key areas to counter antimicrobial resistance issues.
Researchers are using machine learning to identify patterns within high volume genetic data sets. High-throughput sequencing technology has resulted in generation of an increasing amount of microbial data. Machine learning techniques are used to achieve greater depth in the interpretation of genetic information such as how an microbial genes may impact drugs, immunity, and resilience.
The ability of machine learning algorithms to handle multi-dimensional big data has given tremendous power. Moreover, the availability of cloud based hardware with graphics processing units (GPUs) has given tremendous fast computational speed and compatibility with various algorithms. The article explained, how the innovative technologies such as AI & machine learning techniques are used with common sequencing technologies like DNA-seq, ChIP-Seq, RNA-Seq, 16S metagenomics, and small RNA analyses to fight against antimicrobial resistance.
Earlier I have explained the Artificial Intelligence to Combat Antibiotic Resistant Bacteria. There I discussed the common framework for multi-agent deep reinforcement learning models for predicting behavior of bacteria and phages in multi-drug environments. Here, I discussed seven top machine learning projects for antimicrobial resistance. These seven projects are overlapping in nature and used many common tools.
Antibiotic and Antimicrobial Resistance Basics
Understanding the difference between antibiotic and antimicrobial resistance is important. Microbes are living organisms that multiply frequently and spread rapidly. They include bacteria, viruses, fungi and parasites. Antimicrobial resistance (AMR) occurs when microbes such as bacteria, viruses, fungi and parasites change in ways that render the medications used to cure the infections they cause ineffective. Antimicrobial resistance is the broader term for resistance in different types of microorganisms and encompasses resistance to antibacterial, antiviral, antiparasitic and antifungal drugs.
Antibiotic resistant bacteria are bacteria that are not controlled or killed by antibiotics. They are able to survive and even multiply in the presence of an antibiotic. These bacteria currently kill an estimated 700,000 people globally each year – a death toll which could rise to 10 million a year by 2050 if we don’t act [1]. The main difficulty is that the bacteria are changing fast. They changing faster than we can change the drugs in response.
Antimicrobial resistance (AMR or AR) is the ability of a microbe to resist the effects of medication that once could successfully treat the microbe. The term antibiotic resistance (AR or ABR) is a subset of AMR, as it applies only to bacteria becoming resistant to antibiotics. The ability of bacteria and other microorganisms to resist the effects of an antibiotic to which they were once sensitive.
How Microbes Develop Resistance to Drugs
According to the World Health Organization, antimicrobial resistance occurs mainly due to inappropriate use of medicines, for example using antibiotics for viral infections such as cold or flu, or sharing antibiotics. Low-quality medicines, wrong prescriptions and poor infection prevention and control also encourage the development and spread of drug resistance. Lack of government commitment to address these issues, poor surveillance and a diminishing arsenal of tools to diagnose, treat and prevent also hinder the control of antimicrobial drug resistance.
Impact of Antimicrobial Resistance
According to the NIH: National Institute of Allergy and Infectious Diseases, antimicrobial resistance makes it harder to eliminate infections from the body as existing drugs become less effective. As a result, some infectious diseases are now more difficult to treat than they were just a few decades ago. As more microbes become resistant to antimicrobials, the protective value of these medicines is reduced. Overuse and misuse of antimicrobial medicines are among the factors that have contributed to the development of drug-resistant microbes. AMR can lead to the following key issues:
- Many infections being harder to control and staying longer inside the body
- Longer hospital stays, increasing the economic and social costs of infection
- Higher risk of disease spreading
- Greater chance of fatality due to infections
Machine Learning Techniques for Antimicrobial Resistance
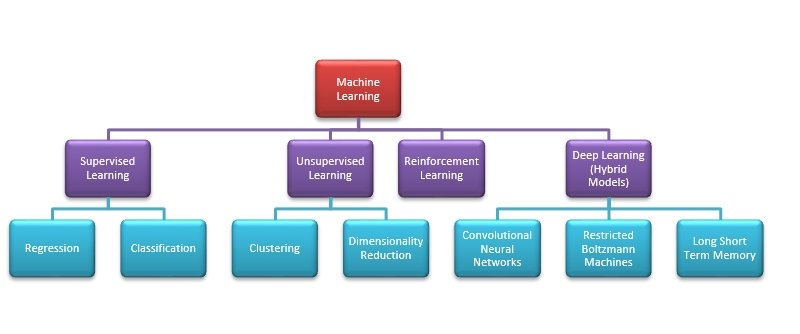
Machine learning techniques can learn from data, without requiring explicit, programmatic instruction. They find patterns from data without the constraints of formulas or even human theories. By learning directly from data, machine-learning techniques can often achieve accuracies not possible with more conventional approaches. One exciting and promising approach now being applied in the antimicrobial field is deep learning, a variation of machine learning that uses neural networks to automatically extract novel features from input data. The availability of vast data of various types antibiotic resistance microbes ensures that there are enough training datasets to build accurate prediction models relating to microbe behaviors and gene expressions. Primarily, there are four approaches to machine learning: supervised learning, unsupervised learning, reinforcement learning and deep learning.
In supervised learning, all data is labeled and the algorithms learn to predict the output from the input data. The supervised learning algorithms includes models like; logistic regression, random forest classification, a boosted decision tree classifier, support vector classification with radial basis function kernel, K-nearest neighbors classification and neural networks. Gradient Boosting is another technique for performing supervised machine learning tasks. XGBoost is particularly popular because it has been the winning algorithm in several recent competitions.
In unsupervised learning models all data is unlabeled and the algorithms learn to inherent structure from the input data. The algorithms discovers relationships between different features on its own. Clustering and dimensionality reduction are the two main uses of unsupervised learning. Principal component analysis (PCA), singular value decomposition (SVD) and k-means clustering are the key algorithms for unsupervised learning models. As microbes have higher data dimensions, so feature dimensionality reduction is also an important part of data processing.
Reinforcement learning is a subset of machine learning, that expands on Markov-decision processes (MDPs), by embedding reward-based feedback into decision outcomes so that an optimal decision approach, termed the policy, can be identified. The vital power of reinforcement learning is that it allows the computer to learn its own approach to obtain a given reward, rather than relying on human behavior as the gold standard.
Deep learning is a subset of machine learning algorithms that allows computational models that are composed of multiple processing layers to learn representations of data with multiple levels of abstraction. These are mostly hybrid models of many hidden layers.
Deep Learning Algorithms
Popular deep learning algorithms includes: Multi-layer perceptron (MLP), Deep Convolutional Neural Networks (CNN), Deep Residual Networks, Capsule Networks, Recurrent Neural Networks, Long Short Term Memory (LSTM) Networks, Deep Autoencoders, Deep Neural SVM, Boltzmann Machines (BM) and Restricted Boltzmann Machines (RBM), Deep Belief Networks, and Recurrent Support Vector Machines.
Recently, the deep convolutional neural networks (DCNNs) have shown astounding performance in object recognition, classification and detection. The basic components of a CNN are stacks of different types of hidden layers (convolutional, activation, pooling, fully-connected, softmax, etc.) that are interconnected and whose weights are trained using the backpropagation algorithm [7]. The training phase of a CNN requires huge numbers of labelled data to avoid the problem of over-fitting; however, once trained, CNNs are capable of producing accurate and generalizable models that achieve state-of-the-art performance in general pattern recognition tasks. Some examples of Deep CNN include AlexNet, GoogLeNet, VGGNet, and ResNet.
Deep Reinforcement Learning (DRL)
The most well-known forms of reinforcement learning algorithms are: Temporal Difference (TD) learning, Actor-Critic, and Q-Learning. The Q-learning functions are used to create and update Q-tables. Deep Reinforcement Learning (DRL) uses of Deep Feedforward Neural Netowrk (FNN) and Recurrent Neural Network (RNN). DQN often uses large deep CNNs for better representation learning. In Deep Q-Learning (DQN), artificial neural network architectures are used for learning. Depending on applications and deep reinforcement learning techniques such as Q-Learning, TD Learning, Partially Observable MDP (POMDP) learning, Actor-Critic Methods of learning, Double DQN, DDQN, Neural Fitted Q Learning, Deep Recurrent Q Network (DQRN) and A3C Algorithms are used in layered structure.
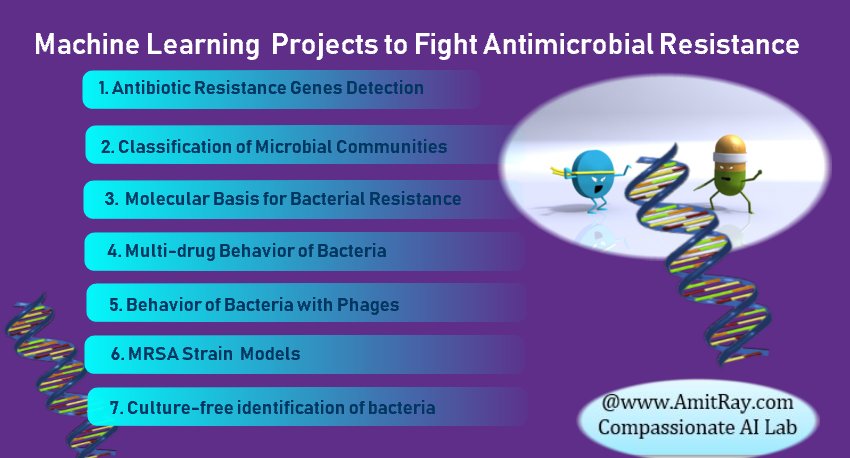
Seven Key Machine Learning Projects for Antimicrobial Resistance
The machine learning application for combating antibiotic resistance bacteria is a new field and growing rapidly. With the development of microbial sequencing in recent years, the microbiome has become increasingly popular in many studies. We have identified seven key projects for antimicrobial resistance. They are; machine learning for antibiotic resistance genes detection, machine learning for classification of microbial communities, machine learning for molecular basis for bacterial resistance, machine learning for multi-drug resistance behavior of bacteria, machine learning for behavior of bacteria with phages, machine learning for MRSA Strains for Hospital-Acquired Infections, machine learning for culture-free identification of bacteria.
In these models, generally, for the bacterial growth model, we assume that the total bacterial population is comprised of drug-susceptible growing cells and drug-insensitive resting cells. The antibacterial effect of the drug is included in the killing rate of the bacteria. By training machine learning classifiers on information about the presence or absence of genes, their sequence variation, and gene expression profiles, we generated predictive models and identified biomarkers of susceptibility or resistance to commonly administered antimicrobial drugs.
The human microbiota consists of about 100 trillion microbial cells, compared with our 10 trillion human cells, and these microbial symbionts contribute many traits to human immunity and biology. Compositional differences between microbial communities residing in various body sites are large, and comparable in size to the differences observed in microbial communities from disparate physical habitats.
In traditional “phenotypic” testing, bacteria are grown in the presence of different concentrations of various antibiotics. Bacteria that do not grow in the presence of a test antibiotic are called ‘susceptible’ and those that do grow are called ‘resistant.’ Today, however, the same information can be generated through newer technology called ‘whole genome sequencing’ (WGS).
With the advent of affordable whole-genome sequencing (WGS) technology, it is now possible to determine and evaluate the entire DNA sequence of a bacterium. By providing definitive genotype information, WGS offers the highest practical resolution for characterizing an individual microbe. Bacteria that have identical resistance patterns caused by different mechanisms can also be differentiated by WGS. Emerging tools such as metagenomics are focusing on applying sequencing to the sample itself, eliminating the need to isolate and sequence bacteria from the sample. Researchers have used WGS analysis extensively to understand the molecular basis and host-ecosystem relationships in infectious diseases and microbiology. This has allowed advances in our understanding of epidemiology, pathogen evolution and virulence determinants to better conduct disease outbreak investigation and assess disease transmission networks.
In the background of WGS and metagenomics, the summary of the seven machine learning applications to antimicrobial resistance projects are discussed below:
1. Antibiotic Resistance Genes Detection
Whole-genome sequence (WGS) of microorganisms has become an important tool for antibiotics resistance screening and, thus, provides rapid identification of antibiotic resistance mechanisms. Sequencing the entire genome is found to be helpful in many antimicrobial applications such as new antibiotics development, diagnostic tests, the management of presently available antibiotics, and clarifying the factors promoting the emergence and resistance of pathogenic bacteria. We have tested DCNNs, Restricted Boltzmann Machines (RBM), Deep Belief Networks algorithms for antibiotic resistance genes detection and the results are impressive.
2. Classification of Microbial Communities
Based on supervised learning techniques microbes can be grouped into various classes based on correlations. Three popular techniques for microbial community classification include convolutional neural networks (CNN), genetic programming (GP), random forests (RF), and logistic regression (LR). Random forests classifier is one of the top performers in microarray analysis. RFs are an extension of bagging, or bootstrap aggregating, in which the final predictions of the model are based on an ensemble of weak predictors trained on bootstrapped samples of the data.
3. Molecular Basis for Bacterial Resistance
Machine learning techniques are used to predict environmental and host phenotypes using microorganisms. One of the major drivers of resistance spreading between bacteria are transposons. This is also called jumping DNA, where the genetic elements that can switch locations in the genome autonomously. When transferred between bacteria, transposons can carry antibiotic resistance genes within them. Use of reinforcement learning to understand the transfer mechanisms in real life is one of our important project. We have noticed, Partially Observable MDP (POMDP) learning models are best fitted for our prototypes testing data.
4. Multi-drug Resistance Behavior of Bacteria
Machine learning techniques to develop the dosing strategies for multi-drug-resistant bacteria is one of the key research area. Multi-drug-resistant bacteria requires aggressive treatment with the limited arsenal of effective, therapeutic antibiotics. Conventional antibiotic combinations have not completely addressed the challenge of treating infections caused by multi-drug-resistant bacteria, especially when incremental and ineffective antibiotic dosing strategies are employed that do not overcome individual mechanisms of resistance. A reinforcement-learning algorithm predicted which patient characteristics and dosing decisions that resulted in the lowest risk of failure to be discharged on the medication. We have noticed, Deep Recurrent Q Network (DQRN) learning algorithms are most suitable for handling multi-drug issues.
5. Behavior of Bacteria with Phages
Phage is a type of virus that infects bacteria. Phage will only kill a bacterium if it matches to the specific strain. Bacteria behave differently when they are exposed to phages. Phage-bacteria interactions are unique in that phage interactions with bacteria can range from predatory, to parasitic. Different types of interactions lead to different evolutionary scenarios for the host population, and different genomic signatures. The use of phages against pathogenic bacteria can be modeled using two different approaches, one passive, the other active. Different bacteria–phage interactions occur depending on the health status and development stage of the human host. We are using various reinforcement learning algorithms to model the bacteria-phage interactions.
6. MRSA Strain Models for Hospital-Acquired Infections
Methicillin-resistant S. aureus (MRSA) in particular has emerged as a widespread cause of both community as well as for hospital-acquired infections. Recently, MRSA was classified by the World Health Organization (WHO) as one of twelve priority pathogens that threaten human health. MRSA biofilm production, the relationship of biofilm production to antibiotic resistance, and front-line techniques to defeat the biofilm-resistance system are the three keys areas for this machine learning project.
Rapid and accurate strain typing of bacteria is vital. This would facilitate epidemiological investigation and infection control in near real time. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry is a reliable strain typing tool. The use of MALDI-TOF MS is currently expanding in clinical laboratories due to its rapid and accurate bacterial identification capabilities. MALDI-TOF spectral analysis has been widely used in clinical microbiology for the identification of bacteria, fungi, and mycobacteria. However, the subtle difference among the spectra is difficult to interpret correctly by visual examination. Moreover, the presence or absence of proteins, and assessment of their expression levels are difficult to discriminate. However, machine learning (ML) methods promises to resolve these issues.
Recently, we have experimented the spectra data with deep convolutional neural networks (DCNNs). The datasets were split into training (65%), validation (15%), and test (15%). Two different DCNNs, AlexNet and GoogLeNet, were used to classify the spectra data. Both untrained and pre-trained networks on ImageNet were used, and augmentation with multiple pre-processing techniques. Ensembles were performed on the best-performing algorithms. The best-performing classifier had an AUC of 0.96, which was an ensemble of the AlexNet and GoogLeNet DCNNs.
7. Machine Learning for Culture-free Identification of Bacteria
To confidently select a narrow-spectrum rather than broad-spectrum antibiotic therapy, the physician requires the pathogen’s antibiotic susceptibility profile. Currently, due to the relatively long time for diagnostic results to be finalized, physicians are often left with no alternative but to employ an empirically defined broad-spectrum antibiotic therapy to secure a patient’s survival. For example, bloodstream infections generally involve extremely low (e.g., <10 colony-forming unit (CFU)/mL) bacterial concentrations, which require a labor-intensive process and as much as 72 hours to yield a diagnosis.
Emerging automated rapid microbiology methods, especially those employing miniaturized microfluidic devices (or lab-on-a-chip systems) and nanotechnologies, offer unique opportunities to combat the crisis of antibiotic resistance. However, along with the allied new technologies, machine learning based culture-free identification of bacteria can reduce the time to 2 hours. We have experimented with XGBoost algorithms.
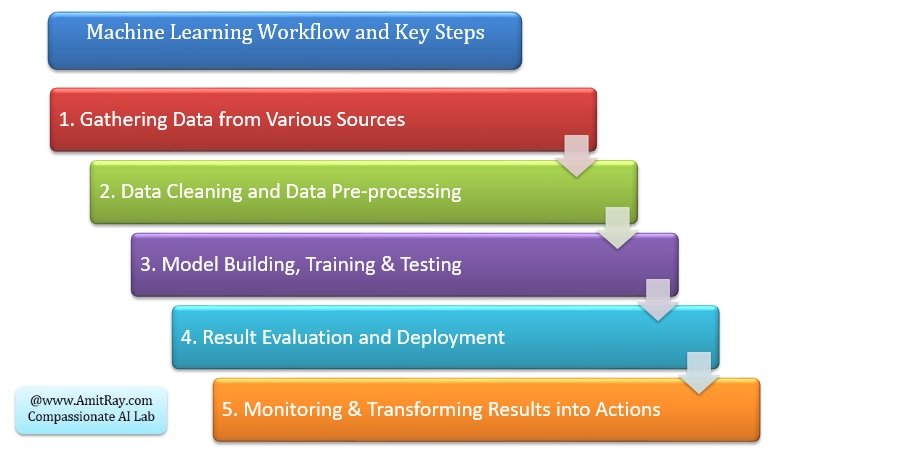
Machine Learning Workflow and Key Steps
The five key steps for the machine learning antimicrobial resistance projects are: gathering data from various sources, data cleaning and data pre-processing, model building training and testing, result evaluation and deployment, monitoring and transforming results into actions.
Key Antimicrobial Resistance Databases for Machine Learning
The Integrated Microbial Genomes (IMG) data warehouse is a leading comprehensive resource devoted specifically to microbes. CARD: The Comprehensive Antibiotic Resistance Database is a rigorously curated collection of characterized, peer-reviewed resistance determinants and associated antibiotics, organized by the Antibiotic Resistance Ontology (ARO) and AMR gene detection models.
GenBank is a free, public collection of all available DNA sequences. It is served by an online platform. Among the existing features, GenBank supplies the DNA’s correspondent protein translation sequences and gives the user the possibility of downloading large sets of records at once.
The freely available PATRIC (Pathosystems Resource Integration Center), platform provides an interface for biologists to discover data and information and conduct comprehensive comparative genomics and other analyses in a one-stop shop. Similarly, PathogenPortal Hub site, EuPathDB for Eukaryotic Pathogens, IRD for Influenza Research Database, ViPR for virus Database and Analysis Resources are full with resources. Microbial Genome Database (MBGD), and the MicrobesOnline resource contains information on thousands of bacterial, archaeal, and fungal genomes. It also provides access to gene expression and fitness data.
Conclusion:
We have discussed seven machine learning projects to fight against antimicrobial resistance microbes, which causing harm and sufferings to the humanity. The objectives of these projects are to prevent microbes from developing resistance to drugs. Machine learning, genomics and multi-omics data integration are the fast-growing emerging technologies to counter antimicrobial resistance problems. While there are great promises, developing large-scale long-term strategies to counter antimicrobial resistance is still an uphill battle.
Deep learning and other machine learning techniques are increasingly showing superior performance in many areas of antimicrobial resistance, which has many complex data. However, there are many obstacles and number of issues remain unsolved for implementing large-scale integrated machine learning systems for antimicrobial resistance. We are eager to embrace machine learning methods as an established tool for antimicrobial resistance prevention and analysis, and we look forward with great anticipation to the new insights that will emerge from these applications.
References:
- Mechanisms of Antibiotic Resistance, Microbiol Spectr. 2016 Apr; 4(2), by Jose M. Munita ,and Cesar A. Arias
- Antibiotic resistance – World Health Organization
- Whole-Genome Sequencing for Detecting Antimicrobial Resistance in Nontyphoidal Salmonella
- Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype
-
Ensemble of convolutional neural networks for bioimage classification