Artificial Intelligence to Combat Antibiotic Resistant Bacteria
Artificial Intelligence to Combat Antibiotic Resistant Bacteria – tools, techniques, models, scopes and challenges are discussed. Antibiotic resistance bacteria is one of the key research area of our Compassionate AI Lab. Dr. Amit Ray explains how artificial intelligence can be used in combating these superbugs. Antibiotic resistance bacteria is becoming world’s biggest health crisis. We discussed here multi-agent deep reinforcement learning models for predicting behavior of bacteria and phages in multi-drug environments. We call this model as DeepCombat.

Antibiotic resistant bacteria are bacteria that are not controlled or killed by antibiotics. They are able to survive and even multiply in the presence of an antibiotic. These bacteria currently kill an estimated 700,000 people globally each year – a death toll which could rise to 10 million a year by 2050 if we don’t act [1]. The main difficulty is that the bacteria are changing fast. They changing faster than we can change the drugs in response.
Artificial intelligence is showing alternative means of fighting these deadly infections and killer bacteria. Multi-drug-resistant bacterial infections annually result in millions of hospital days, billions in healthcare costs, and, most importantly, thousands of lives lost. Artificial Intelligence for healthcare is progressing at an exponential rate. We are evaluating here, the role of artificial intelligence in fighting these superbugs. Especially, the use of AI for intelligent Phage therapy.
Common Antibiotics
Antibiotics are chemical compounds used to kill bacteria or inhibit the growth of infectious organisms (like Bacteria, protozoa). Alexander Fleming’s discovery of penicillin in 1928, which has rightfully been described as a “turning point in history”. After that, humans use hundreds of thousands of tons of antibiotics per year for medical, veterinary, and agricultural purposes. There are 5 generations of antibiotics. They are classified depending on their spectrum and bacteria killing abilities.
The 14 common antibiotics are; amikacin, amoxicillin/clavulanic acid, ampicillin, cephalexin, ceftiofur, cefuroxime, ciprofloxacin, clindamycin, cotrimoxazole, enrofloxacin, gentamicin, norfloxacin, ofloxacin and tetracycline.
Classification of Bacteria:
Bacteria are classified into two groups based on the structural differences in their cell walls: gram-negative and gram-positive bacteria.
Gram-negative bacteria have a thin membrane, which is nearly “bulletproof.” Gram-negative bacteria’s cell membrane is thin but difficult to penetrate. Because of this nearly “bulletproof” membrane, they are often resistant to antibiotics and other antibacterial interventions. Examples of Gram-negative bacteria include cholera, gonorrhea, Escherichia coli (E. coli), Klebsiella pneumoniae, and Acinetobacter baumannii. These bacterial pathogens grow increasingly resistant to current antibiotics.
Gram-positive bacteria have a big, thick membrane. Gram-positive bacteria have a big, thick membrane but easy to penetrate. Examples of Gram-positive bacteria include Streptococcus, Staphylococcus, and Clostridium botulinum (botulism toxin).
How Bacteria Develop Resistance to Antibiotics
Every time a person takes antibiotics, sensitive bacteria are killed, but resistant bacteria may be left to grow and multiply. Repeated and improper uses of antibiotics are primary causes of the increase in drug-resistant bacteria. Normally, bacteria become resistant in three ways: 1) by a genetic mutation or 2) by acquiring resistance from another bacterium 3) by changing in membrane permeability to antibiotics.
Gene mutation of Bacteria
A gene mutation is a permanent alteration in the DNA sequence that makes up a gene. Mutations lead to variations in genes. Variations are also very important in evolution. Without variations evolution would not be possible, and changes in any part of the environment affecting the organism could make the organism become extinct.
Creation of Biofilm Barrier
Over time, bacteria have evolved sophisticated mechanisms of drug resistance to avoid killing by antibiotics. Creation of biofilm barrier is one such mechanism. The biofilm provides advantages to bacteria, such as enhanced resistance towards environmental stresses, including antibiotic challenge. Often extracellular DNA (eDNA) forms part of biofilm matrices. Extracellular DNA can also act as a shield.
Acquiring Resistance from another Bacteria
Bacteria can acquire antibiotic resistance genes from other bacteria. Bacteria can transfer genetic material, including genes encoding resistance to antibiotics by undergoing a simple mating process called “conjugation”.
Multi-agent AI System to Combat Antibiotic Resistance Bacteria
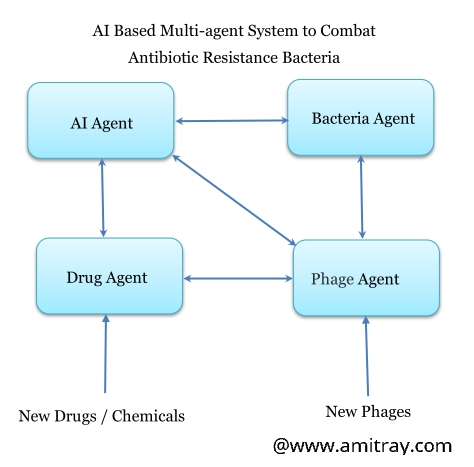 We have experimented various artificial intelligence based multi-agent deep reinforcement learning models with modern biotechnology to analyse the behavior of the bacteria and the phages. We call this system as DeepCombat.
We have experimented various artificial intelligence based multi-agent deep reinforcement learning models with modern biotechnology to analyse the behavior of the bacteria and the phages. We call this system as DeepCombat.
Agent-based AI technology is used to model and simulate the dynamics of the system. The fundamental characteristic of the agents are their ability to sense their environment, react in an autonomous manner and fulfill their long-term and short-term objectives.
The schematic diagram of the DeepCombat model is shown in the diagram. It has four agents: AI agent, Bacteria Agent, Drug Agent, and Phage Agent. Each agent have their separate value functions and policy functions. The AI agent is the master and having the global value function. The system works in a master-slave based multi-agent system. The AI agent works as the master and the other three acts as slave.
Intelligent AI Agent:
The system simulates the complex intelligent battle between bacteria and the phage. Each type of phage infects a specific type of bacteria. Finding the right type of phage which can defeat the bacteria is the objective of the value function of the AI agent. Here the AI agent behave like a living cells. They are intelligent like nano-machines that can detect and process many signals and respond to them.
Bacteria Agent:
Scientists observed that a bacterial colony loosely mimics a biological neural network. The bacteria can take inputs in form of chemical signals, process them and then produce output chemicals to signal other bacteria in the colony. We can predict behavior of bacteria by reading the content of the gene. Bacteria are capable of sensing and interacting with variety of environmental signals: amino acids, sugars, oxygen, pH, temperature, light, environmental DNA, and other microbes —even physical objects such as tiny, chemically inert beads. Moreover, bacteria behave differently when they are continuously exposed to antibiotics, and the rate and extent to which resistance develops are strongly dependent on the type of bacterial species and type of drug. Models includes natural biofilm development cycle.
Intelligent Drug Agent:
We have simulated multi-drug environments with the intelligent Drug agent. We have simulated the relationships between antibiotic concentration, bactericidal activity in dynamic environment with deep convolutional neural network. This technique automatically learn the best feature representation from the training data. Interactions between drugs are also modeled. In some cases we find that antibiotic resistance to some synergistic combinations evolves faster than resistance to individual drugs.
Phage Agent
Phage is a type of virus that infects bacteria. Phage will only kill a bacterium if it matches to the specific strain. Bacteria behave differently when they are exposed to phages. Phage-bacteria interactions are unique in that phage interactions with bacteria can range from predatory, to parasitic. Different types of interactions lead to different evolutionary scenarios for the host population, and different genomic signatures. The use of phages against pathogenic bacteria can be modeled using two different approaches, one passive, the other active. Different bacteria–phage interactions occur depending on the health status and development stage of the human host.
Minimal Bacterial Gene Set
A minimal gene set (MGS) is defined as the smallest possible gene set necessary and sufficient to maintain a living organism. Computational analysis has extensively used to try to get closer to the minimal gene set to represent a bacteria. The gene set must contain all the information to allow the cell to perform many essential (housekeeping) functions that give the cell the ability to maintain metabolic homeostasis, reproduce, and evolve, the three main properties of living cells. Researchers have designed and synthesized a minimal bacterial genome, containing only the genes necessary for life, and consisting of just 473 genes. However, the following 38 genes associated with pathogenicity are considered for the initial AI model: crl, ireA, cnf1/2, tia, sat, fyuA, mat, sfa/focCD, malX, afa/drab, neuC, iha, hrlA, fimC, pic, hlyA, kpsMTII, sitDep, ompA, iroN1, gimB, sitD, tratT, ibeA, chuA, vat, tsh, iucD, cvi/cva, papC, irp2, iss, EAST1, felA, iutA, cvaC, papG, fimH.
AI for predicting behaviors of bacteria and phages
Predicting phenotype of bacteria and phages from genotype with deep reinforcement learning algorithms is comparatively easy. The genotype is the set of genes in the DNA which is responsible for a particular trait. The phenotype is the physical expression, or characteristics, of that trait. Genotype is what makes the trait – the information within a gene. Phenotype is what you see – the visible or observable expression of the results of genes. DNA sequencing. The use of artificial intelligence makes finding the right phage much easier, turning the strategy into a more practical treatment option for antibiotic resistance bacteria.
Conclusion:
We have discussed here the architecture of the artificial intelligence based system to combat antibiotic resistance bacteria. This is a complex multi-agent master-slave hierarchical deep reinforcement learning system. In this model there are several issues to be resolved. Tuning the models and determining the parameter of the reinforcement learning and CNN of the AI based multi-agent antibiotic resistance bacteria model is a critical issue. However, the initial experimental results show that the proposed approach is promising and effective.
Source
Compassionate Artificial Superintelligene AI-5.0, Dr. Amit Ray, 2018
References:
- Mechanisms of Antibiotic Resistance, Microbiol Spectr. 2016 Apr; 4(2), by Jose M. Munita ,and Cesar A. Arias
- The cognitive cell: bacterial behavior reconsidered, Front Microbiol. 2015; 6: 264. by Pamela Lyon
- Dependency-based convolutional neural network for drug-drug interaction extraction, 2017 IEEE International Conference on, pp. 5-12, 2017 by Ramakanth Kavuluru, Anthony Rios, Tung Tran.
-
Antibiotic Discovery: Combatting Bacterial Resistance in Cells and in Biofilm, Communities, Molecules 2015, 20(4), 5286-5298 by Anahit Penesyan, Michael Gillings and Ian T. Paulsen
-
A novel proposal of a simplified bacterial gene set and the neo-construction of a general minimized metabolic network, Nature, Scientific Reports volume 6, Article number: 35082 (2016) by Yuan-Nong Ye, Bin-Guang Ma, Chuan Dong, Hong Zhang, Ling-Ling Chen & Feng-Biao Guo.
